Our Vision
Our vision is to realize and promote cleaner waters that sustain biodiversity in both natural and human-impacted habitats.
“Clean, unpolluted waters host more biodiversity than disturbed ones.” — GIGA New River Research Team
Funding Provided by: 
The Project
The non-profit Global Invertebrate Genomics Alliance (GIGA) has been awarded a 2023 Climate Change and Human Health Seed Grant by the Burroughs Wellcome Fund. The primary aim of our project is to determine the current, baseline health of the New River of Fort Lauderdale where it enters the Sistrunk community in South Florida. This will be accomplished using state-of-the-art molecular microbiology methods. A second goal of this project is to form new alliances among diverse communities – sciences, academia, social sciences, and resident stakeholders in the context of conserving biodiversity and climate change.
New River Our Home Project – Community Report (6/6/24)
Our Methods
A baseline of public climate literacy within the Sistrunk neighborhood will be assessed through a pre-test survey instrument at the initial citizen science event introducing the project and working to gain community participation and interest. Biological sampling results will be shared with the community through a series of inter-generational public education and engagement activities including citizen science, and science, technology, engineering, art, and mathematics (STEAM) workshops to bolster interest, knowledge and understanding of how humans impact our environment, historically, contemporarily and the actions that are needed now in order to ensure a viable future for our community and our species. The intersections of biology, health, human impacts on the environment and climate change will be key factors of discussion and deliberation. Factsheets/ checklists will be designed to distribute to participating households as a reminder and reference to information exchanged about how to reduce pollution and climate related human health disparities and what steps can help to increase the quality of our riverine ecosystem and our lives and longevity as residents within the New River biome. A post-test survey instrument delivered at the conclusion of the second biannual citizen science workshop event will assess the extent to which the project was successful in increasing the climate literacy of Sistrunk community residents.

Personnel
- Prof. Jose V. Lopez, Principal Investigator
- Dr. Tara Chadwick, Co-PI
- Dr. George Duncan
- Dr. Alcee Hastings II
- Stacy Brown, Graduate Research Assistant
Study Sites
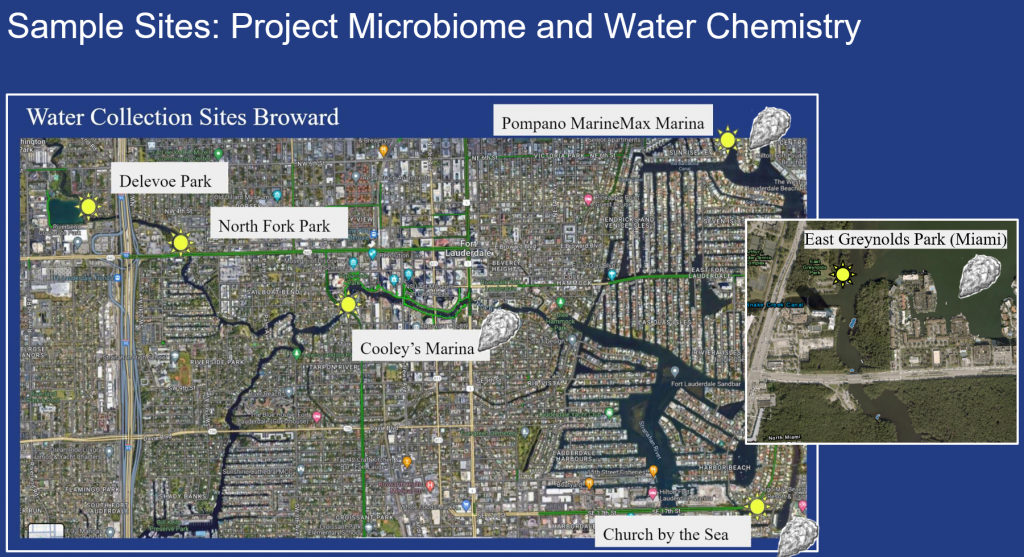
Buroughs Wellcome Fund supported phase one of the project which included conducting baseline water quality assessments at sites (not yet mapped but reported to have oyster populations) along the New River, Intracoastal and Pompano Beach in Broward and in East Greynolds Park in Miami. (3) sites, Delevoe Park, North Fork Riverfront Park, and Cooley’s Marina are part of the Sistrunk Sites and (3) sites, Pompano Marina, Church by the Sea and East Greynolds Park were confirmed oyster sites.
Results
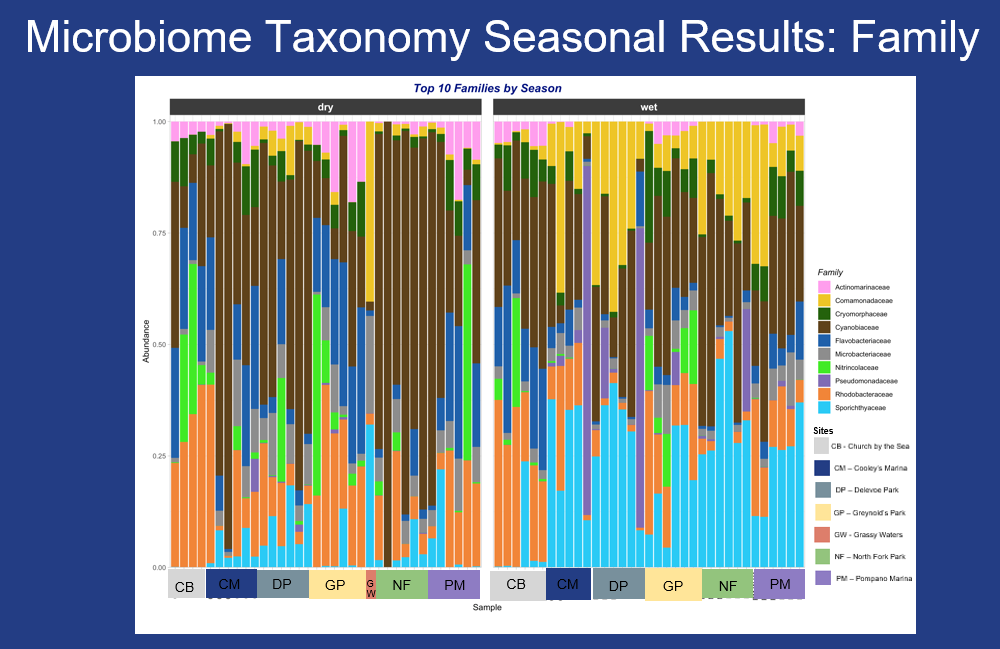
Health concerns are voiced by residents who use the waterways frequently, this study indicated a higher abundance of Pseudomonadaceae along the New River sites and both Sporicthyaceae and Comamonadaceae across the sites during the wet season. A higher abundance of Cyanobiaceae, Rhodobacteraceae and Actinomarinaceae were present during the dry season.
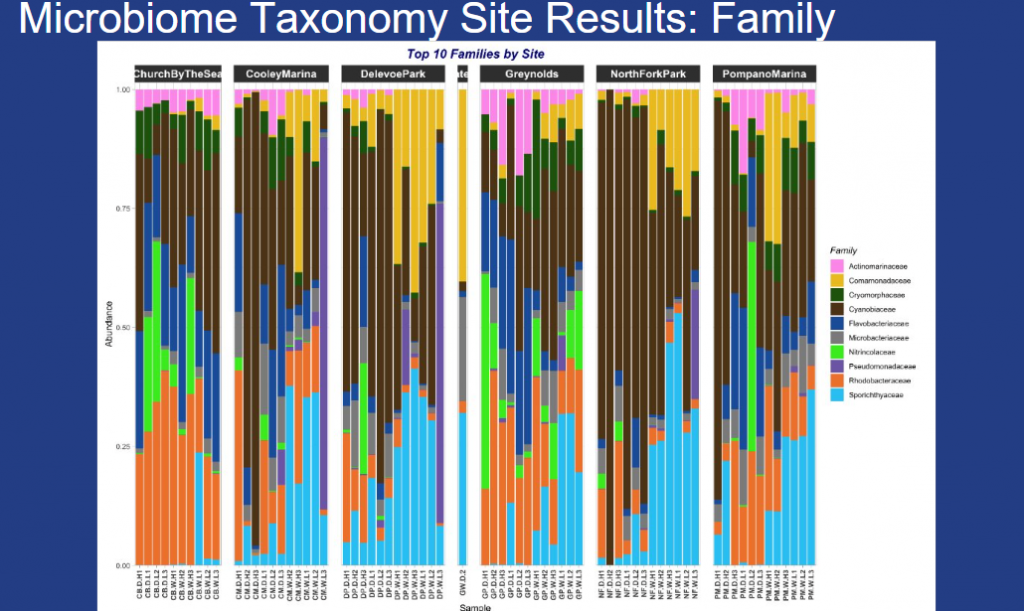
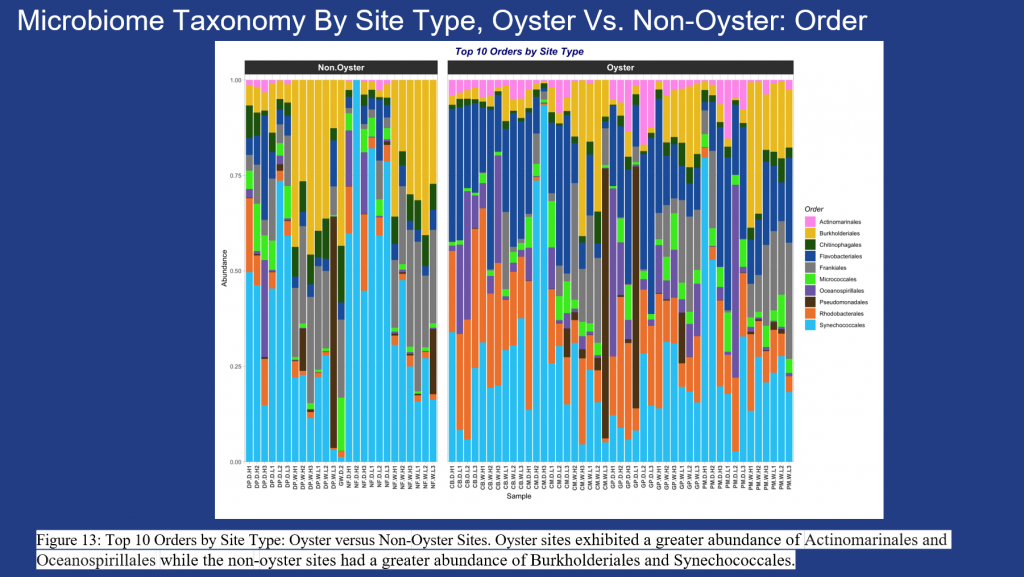
Microbial abundance varied across the study sites with Cyanobiaceae and Comamonadaceae more common along the Sistrunk Freshwater sites. Actinomarinaceae, Rhodobacteraceae, Flavobacteriaceae and Nitrincolaceae were more abundant in the saltwater sites.
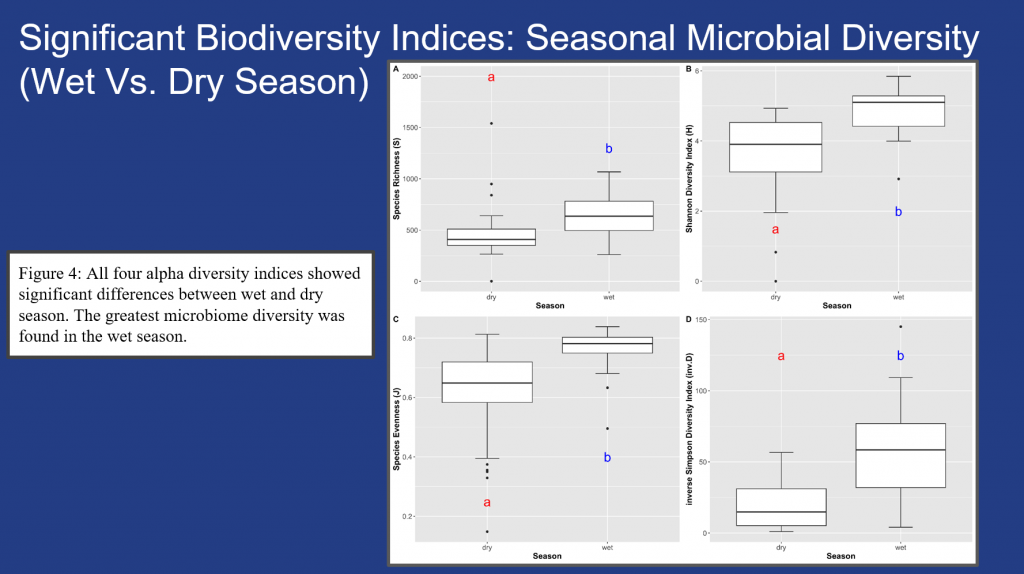
Significant seasonal shifts within the microbiome biodiversity were confirmed among the sites across alpha diversity indices and the wet season had more significant microbial diversity than the dry season across all the samples collected (Fig. 4).
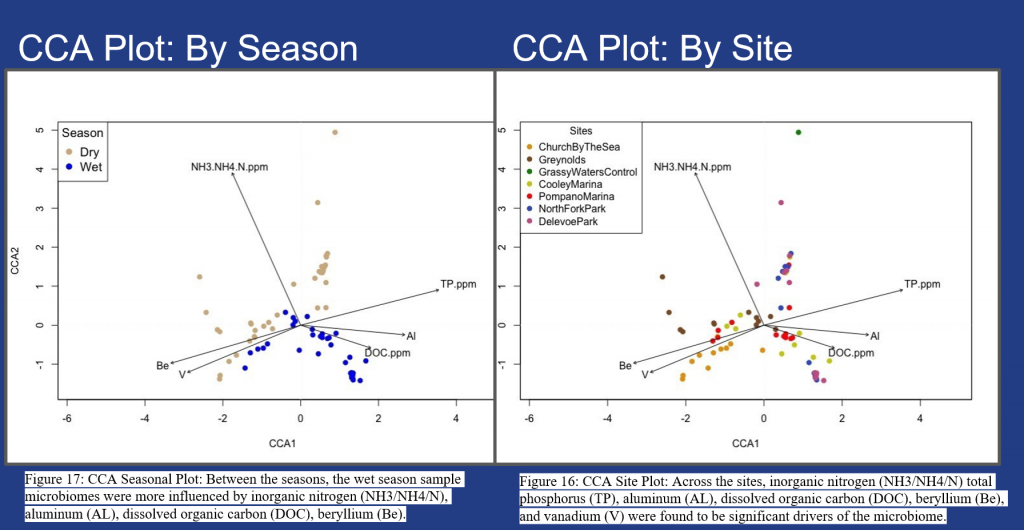
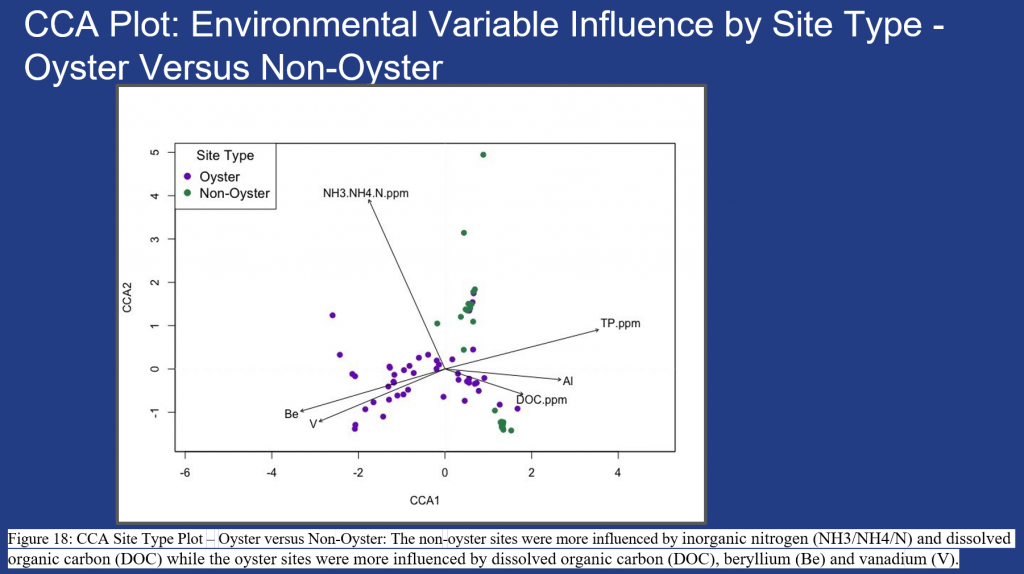
Figure 16 – 18, show seasonal and site specific environmental variables driving the microbiome at the study sites. Significant seasonal environmental drivers include inorganic nitrogen, dissolved organic carbon and total phosphorus and metals beryllium (Be), aluminium (AL) and vanadium (V), Fig. 16. Significant differences are shown with greater clustering of samples during the wet season (in blue) and dry season (in tan) within Fig. 17. Additional research is needed to determine the impact on fish and invertebrate species due to the metals (Be, AL & V) found within the sampled sites, Fig. 18.
Community Engagement

Citizen science, community input and community education intersect with the more research driven components of this study such as the microbial and water quality analysis. Over time, as oysters and other filter feeding species have been removed from the habitat with greater frequency, water quality has suffered. The community of focus in the New River: Our Home project, Sistrunk, has experienced water quality issues that have impacted the community’s ability to swim, fish and utilize the New River corridor. The study sites of Delevoe Park, North Fork River Park, and Cooley’s Marina are no exception with residents reporting fewer days with clear waterways, increased pollution and decreased fish and invertebrate biodiversity.
Several meetings were held to discuss the results of this study with the community to gather information as to how they, as citizen scientists, would like to contribute to future restoration of the Sistrunk sites. Surveys were collected at (2) public outreach days, January 2023 Dr. Martin Luther King Jr. Sistrunk Parade and Delevoe Park Waterway Clean-up. Per Dr. Tara Chadwick, Co-PI and community member, “Over the first 6 months of the project, 300 community members were educated on the links between environmental and human health. Of these, 40 completed a brief survey indicating an interest in addressing local water quality, biodiversity and health issues.” A diverse segment of the population was reached which included various age demographics. Survey questions included a range of environmental knowledge to use impact related. Residents were interested and engaged in the efforts. Input from residents, businesses and government officials was considered and included in this study.
Project information sharing included water chemistry data publication on publicly available websites and the inclusion of raw data in genetic databases such as NCBI.
As oysters are filter feeders, an increase in oyster populations within South Florida could help to reduce the high abundance of algal species within the waterways. This study supports that oyster populations are low within the regions studied and oyster restoration in South Florida could be considered as part of a natural solution implemented within local water quality improvement projects.
Resources and References
- https://www.broward.org/NaturalResources/Lab/Documents/pub_newriver_1.pdf
- https://floridadep.gov/dear/water-quality-evaluation-tmdl/documents/c-14-cypress-creek-canal-wbid-3270-c-13-west-middle (link to downloadable pdf)
- https://www.fortlauderdale.gov/home/showdocument?id=884 (2006)
- http://edr.state.fl.us/content/natural-resources/LandandWaterAnnualAssessment_2022Edition_Volume2.pdf (2022)
- Sea Level Solutions Day Citizen Science Project with FIU & City of Miami https://environment.fiu.edu/what-we-study/projects/slsc-citizen-science/
- SDG 4 priorities for urban coastal climate adaptation https://sdg.iisd.org/commentary/guest-articles/ocean-climate-platform-advocates-for-adapting-cities-to-sea-level-rise/ accessed 11.25.23 2:54 am


